|
 |
|||||||||||
|
|
|||||||||||
|
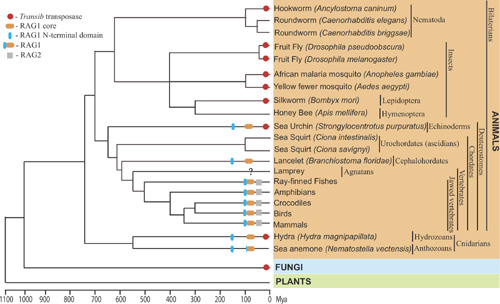
The Revenge of Calvin and HobbesBy Andrea BottaroPosted June 5, 2005 In "Darwin's Black Box" (DBB), ID's arch-biochemist Behe glibly labeled evolutionary hypotheses for the origin of "irreducibly complex" systems as "hops into the box of Calvin and Hobbes" (for those who don't know what the heck this refers to, go here to learn about Calvin and Hobbes, and here for info on their box, or even better go spend some time here, and come back tomorrow). This overconfidence has come back to haunt him as more and more evidence accumulated in support of the evolutionary origin of his various IC systems, from the flagellum to the complement and clotting cascades. The topic where the idea of unevolvability of IC systems has probably taken the most beating is the vertebrate adaptive immune system, where not only evidence for evolution has accumulated at a steady pace, but even more embarrassingly for Behe, it has developed exactly along the lines predicted by those "Calvin and Hobbes jumps" he originally dismissed. A recent paper in the journal PLoS Biology [1] is the latest turn in the death spiral of irreducible complexity of the immune system, and I think provides a good opportunity to take a look at how science works, as opposed to ID navel-gazing. Let me start with a brief description of the issue. Basically, almost every organism faces the problem of pathogens and parasites, and most of them solve it, in broad terms, using molecular detection systems that can discriminate "self" from "non-self", allowing the elimination of the latter. Of course, the problem of discriminating you from them is that there is only one you, and an almost infinite variety of them -- so any effective discrimination system has to be flexible enough to recognize very many different forms of non-self, and able to do so in a parsimonious way in term of use of molecular components (no organism can possibly hope to have a genome large enough to encode even one single specific detector molecule for every possible pathogen). In evolutionary terms, any successful immune system has therefore to satisfactorily juggle, among other things, these two contrasting selective pressures: diversity of effective target vs metabolic/genetic parsimony. Jawed vertebrates (which include cartilaginous and bony fish, amphibians, reptiles, birds and mammals) happen to have hit on a solution that is, frankly, way cool. (And I am not saying it just because I work on this.) In all jawed vertebrates, the adaptive immune system "detectors" (receptors), are encoded not by single, stable genes, but by families of gene segments that change their conformation on DNA (rearrange) during the development of immune cells. By randomly joining one segment from each of the different families (called "V", "D" or "J", each containing from a few to a few hundred members) into a single coding sequence, the immune system can generate many thousands of different genetic combinations, each encoding a different receptor capable of recognizing a different molecular target. The proteins that mediate this DNA rearrangement process ("VDJ recombination") are called RAG1 and RAG2, and they act on specific DNA sequences, recombination signal sequences, or "RSSs", which flank the rearranging V, D and J segments. (For those who want to know more, Matt Inlay's excellent summary, especially its "IC system II" section, serves as a very good primer). Those of you who are used to the ID approach on science, i.e. giving up on it, can probably already see where the problem lies: this is a complex system of functionally inter-related components that, looked at superficially, simply cannot work in isolation. Behe was absolutely certain of this in 1996: In the absence of the machine [RAG1/RAG2], the parts [V, D and J gene segments] never get cut out and joined. In the absence of the signals [RSSs], it's like expecting a machine that's randomly cutting paper to make a paper doll. And, of course, in the absence of the message for the antibody itself, the other components would be pointless. Now, in evolutionary terms the obvious question to ask is indeed what function could any precursor of this system have had, before the evolution of the adaptive immune system. Some ideas were already around at the time of DBB's publication, and had been for a while. Already in 1979, Sakano, Tonegawa (who would later win a Nobel Prize for his discovery of VDJ recombination) and colleagues identified the RSSs and noticed that they shared features with the recombination sequences of certain mobile DNA elements called transposons [2]. Transposons are odd fellows in the DNA world, who spend their time physically hopping from genomic site to genomic site, and replicating themselves, pretty much as "molecular parasites". They do this via a number of mechanisms, but the kind we are interested here are a class of DNA transposons which carry within their own sequences genes encoding the necessary enzymes ("transposases") for cutting themselves off the genomic DNA ("excision"), and re-inserting somewhere else ("integration"). At the very end of each transposon element is a characteristic sequence, which is recognized by the specific transposase (I am sure you are already seeing the parallel with VDJ recombination). A decade after the discovery of VDJ recombination the responsible enzymes, RAG1 and RAG2, were identified, and lo and behold their genes had a funny look about them: just like transposases, they were almost devoid of introns, and mapped right next to each other in the genome (transposons need to "travel light", and cannot carry excess DNA as they hop around). This is when David Baltimore, in whose lab the RAG genes were discovered, and others wrote the review in the Proceedings of the National Academy of Sciences USA [3] that was mocked by Behe as proposing a "hop in the box of Calvin and Hobbes" for openly stating the transposon hypothesis: that RAGs/RSSs were the remnants of some sort of transposon system that integrated itself into a non-rearranging immune receptor, and became "enslaved" to it, causing the integrated portion to "pop out" whenever the gene became active, and in so doing generated useful diversity for immune target recognition. To be fair, at the time the hypothesis was indeed quite a stretch, but still, a stretch that made some specific predictions. No sooner had Behe's words been put to print, those predictions started coming true. What follows is a short timeline, with the major milestones: 1996: - DBB published. In it, Behe says: ... the complexity of the [VDJ recombination] system dooms all Darwinian explanations to frustration. [my emphasis] - In the same year, the Gellert lab, which had developed a system to study VDJ recombination in a test tube with purified proteins, discovers a striking similarity between the RAG-mediated reaction and that of known transposases and integrases: both proceed through a characteristic intermediate in which the DNA takes an unusual hairpin-like shape [4]. This was really a breakthrough finding, the first solid piece of evidence in favor of the transposon hypothesis. But are RAGs actually a transposase? 1998: The Gellert and Schatz lab independently discover that RAGs can, under certain in vitro conditions, mediate actual transposition reactions by inserting cleaved DNA ends containing RSSs into doubleanded target DNA. This is the other side of the transposon "life cycle", the insertion phase. In other words, although the RAGs' physiological activity requires them only to cut DNA out of the genome, they bear, buried inside their structure, the ability to insert RSSs into DNA, a sort of molecular vestigial structure. This makes sense if RAGs are indeed an evolved transposase, but is harder to justify from a design perspective, because transposition events, by potentially disrupting genes, can actually be quite deleterious, for instance by causing cancer. 2000: Indirect evidence is identified that suggests that transposition reactions mediated by RAGs can occur not just in vitro, but within mammalian cells [7]. 2003: Direct evidence of RAG-mediated transposition in yeast and mammalian cells is uncovered [8, 9]. In other words, RAGs are a transposase in eukaryotic cells. 2004: Molecular evidence uncovers a class of transposons called hAT that use a transposition mechanisms essentially identical to that used by RAG proteins, and, in addition, that their enzymes share some basic similarity with RAGs in their active site [10]. (This paper was discussed by Matt here on PT a few months ago) 2005: RAGs find their long-lost family of transposases [1]. In this paper, Kapitonov and Jurka take a fully evolutionary, biocomputational approach to figure out where RAGs may have come from. Let's look at what they did, and how. They started from the observation of a low, borderline significant sequence similarity between a portion of RAG1 and certain transposases of the Transib family. They applied then a different algorithm for protein similarity searches, which uses information from a similarity search to "hone" successive iterations of the same search, by assigning position-specific scores to amino acid residues. In other words, it searches for "deep" homologies that are reflected by the presence of specific sequence motifs within proteins that may otherwise have diverged significantly (and thus yield poor scores at a direct alignment). What they found was that 10 motifs were very highly conserved between RAG1 proteins from various species and Transib transposases. Figure 1 below shows the alignment of these motifs, where every letter corresponds to an amino acid, and color patterns indicate amino acids which have similar physico-chemical properties (and can therefore often replace each other without much disruption of structure and function). The similarities are very statistically significant. Figure 1 -- Alignment of conserved regions in Transib transposases and RAG1 (Click on the figure to see a larger version from the original paper) They next compared the sequence of the RSSs to those of known Transib transposon signal sequences (terminal inverted repeats, or TIRs), and they found another striking correlation: all the positions that are strongly conserved in TIRs are also strongly conserved in RSSs. This is shown in Figure 2. Figure 2 -- Alignment of TIRs and RSSs. Panel A shows a graph of the nucleotide sequence conservation (with 1.0 = absolutely conserved) at different positions in a large panel of TIR families (sequences shown in panel B). Under the graph are the "consensus" TIR sequence (representing the most common nucleotide at each position), aligned with the consensus RSS, which consists of a 7-nucleotide sequence, followed by a spacer, and another conserved 9-nucleotide sequence. Boxed nucleotides are those that show highest conservation, and are absolutely required for the mechanisms. Panel C shows that not only are the crucial positions in the sequences conserved, but their overall structure also is. Like RSSs, which require "spacer" elements of either 12 or 23 nucleotides to pair for efficient rearrangement, so do the TIRs at the ends of each transposon have different and specified length, which correspond to a 12- or 23 nucleotide distance between the conserved sequences (the reason for these numbers is that each turn of the DNA double helix is about 11-12 bp, so sequences 12 or 23 bp apart will be on the same side of the helix, one or two turns apart, and simultaneously accessible to recognition by any binding factor). What this means is that a simple system exists, with both a RAG1-like gene and RSSs, as an independent functional unit: what we would expect for a direct, "reduced" predecessor to the supposedly irreducible VDJ recombinase system. But there's more: while extending their search to the genome databases from various organisms, Kapitonov and Jurka found a number of other RAG1 homologues in various organisms, including some in which the similarity extended beyond the protein "core" they had originally search for, all the way to the so-called N-terminal region of the protein. There is therefore a family of close RAG1-related proteins in various organisms. The distribution of the various homologues in different lineages is shown below. Figure 3: RAG-like proteins and Transib transposons in various organisms. Red circles represent Transib transposons, orange and blue ellipses RAG1 core and N-terminus homologues, and gray rectangles RAG2 proteins. Note that these new RAG1-related proteins are not known to be associated with any Transib-like transposons. Some are clearly pseudogenes, and the function of others is unknown. The overall picture that emerges is that of a complex, diffuse and diverse family, which has accompanied metazoan evolution for a while, with multiple instances of horizontal gene transfer (quite common for mobile DNA elements) and of independent "adoption" by the host genomes of family members. Exactly the picture which one would predict would facilitate the occurrence of random integration of a transposable element within a primordial antigen receptor gene, causing junctional diversification (that is, protein variation at the excision site), and therefore an increase in target binding ability: the "transposon hypothesis" for adaptive immune system evolution. Which brings me to the last item in the story. At the time Behe wrote, no known potential precursor of the immune system receptors existed outside jawed vertebrates. Many proteins belong to the same structural family of antigen receptors, but none carried the same exact sequence hallmarks. That has changed too: at least 3 protein families have been now identified in protochordates and jawless vertebrates which have non-rearranging V-like segments of the same kind of antigen receptors [11, 12, 13]. They show presence of multiple, closely related members, suggesting that selective pressure exists for their diversification, and some may even be involved in "innate" immune responses. Although it is almost impossible to say whether any of these proteins is in fact the direct descendant of the ancestral receptor of the adaptive immune system, their existence suggests a rich evolutionary history of non-rearranging immune receptors predating VDJ recombination adaptive immunity. Let's summarize: where once Behe saw an "irreducibly complex" system made of a) a receptor gene, b) a RAG recombinase, and c) RSSs, we now know that a) whole families of non-rearranging receptors and b) a whole family of functional RAG1 homologues acting on c) RSS-like sequences already existed before the emergence of the vertebrate adaptive immune system. Exactly what we would expect to see if the adaptive immune system did arise via an evolutionary process, as opposed to poof into existence in its complete form. So, what next? Well, for one, we still don't know where RAG2 came from. So far, no RAG2-like genes have not been found, inside or outside transposons. However, the lack of introns and chromosomal location of RAG2, right next to RAG1, are too strong a hint to dismiss, so I think the prediction still remains that a RAG2 ancestor will be found in association with a mobile DNA element, along with a RAG1-like transposase. In the context of VDJ recombination, RAG2 seems to play mostly a regulatory role, so it would not be surprising if its ancestor did something similar. However, it is possible that considerable sequence divergence may have occurred for this protein, since mechanisms for transposon regulation may be significantly different from those required for VDJ regulation. Thankfully, much work remains to be done -- that's what scientists are for. Is Behe going to concede that evolutionary models for the origin of VDJ recombination are gaining more and more support by the day? Probably not, frankly. No matter how many predictions get verified, how many plausible precursors are identified, Behe and the ID advocates will retreat further and further into impossible demands, such as asking for mutation-by-mutation accounts of specific evolutionary pathways, as if one could meaningfully recreate in the lab the precise evolutionary conditions which some mud-dwelling lamprey-like critter experienced some time in the Cambrian. Too much has been invested by ID advocates in the "irreducibly complexity" concept for them to recognize its significance (assuming it ever had any, given its recurrent reformulations) has essentially collapsed. For the rest of us, the lesson to be learned is that even wild hypotheses, if rational, consistent with available evidence, predictive and testable, are worth considering and pursuing. Behe said: We can look high or we can look low, the result is the same. The scientific literature has no answer to the question of the origin of the immune system.Yet the answer was there all along, in the only place where Behe refused to look: in the box of Calvin and Hobbes. AcknowledgementsThanks to Matt Inlay and the rest of the PT crew for info, comments and suggestions. References1. Kapitonov VV, Jurka J. RAG1 Core and V(D)J Recombination Signal Sequences Were Derived from Transib Transposons. PLoS Biol. 2005;3: e181 [Epub ahead of print] 2. Sakano H, Huppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979; 280: 288-94. 3. Bartl S, Baltimore D, Weissman IL. Molecular evolution of the vertebrate immune system. Proc Natl Acad Sci U S A. 1994; 91: 10769-70. 4. van Gent DC, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996; 271: 1592-4. 5. Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998; 94: 463-70. 6. Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998; 394: 744-51. 7. Vaandrager JW, Schuuring E, Philippo K, Kluin PM. V(D)J recombinase-mediated transposition of the BCL2 gene to the IGH locus in follicular lymphoma. Blood. 2000; 96: 1947-52. 8. Clatworthy AE, Valencia MA, Haber JE, Oettinger MA. V(D)J recombination and RAG-mediated transposition in yeast. Mol Cell. 2003; 12: 489-99. 9. Messier TL, O'Neill JP, Hou SM, Nicklas JA, Finette BA. In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. EMBO J. 2003; 22: 1381-8. 10. Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004; 432: 995-1001. 11. Cannon JP, Haire RN, Litman GW. Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nat Immunol. 2002; 3: 1200-7. 12. Cannon JP, Haire RN, Pancer Z, Mueller MG, Skapura D, Cooper MD, Litman GW. Variable domains and a VpreB-like molecule are present in a jawless vertebrate. Immunogenetics. 2005; 56: 924-9. 13. Suzuki T, Shin-I T, Fujiyama A, Kohara Y, Kasahara M. Hagfish leukocytes express a paired receptor family with a variable domain resembling those of antigen receptors. J Immunol. 2005; 174: 2885-91. Originally posted at The Panda's Thumb. Behe's Meaningless ComplexityBy Andrea BottaroIn the Inferno, Dante tells the story of Count Ugolino della Gherardesca (don't even try to pronounce it, unless you are Italian). Count Ugolino was locked up in a tower with his sons, without food or water, by his Pisan political enemies, whom he had betrayed. To survive, he ate his own children (he died anyway, and got to spend eternity stuck in a frozen lake, gnawing at his incarcerator's skull). Michael Behe also had to face Ugolino's choice: starving for support for ID, he was forced to eat his own brain-child, "irreducible complexity" (IC). The meal was fully consumed in Behe's response to my "The Revenge of Calvin and Hobbes" post. Dr. Behe claims that the only evidence that would convince him of the evolution of an IC system consists not only of a complete step-by-step list of mutations, ... but also a detailed account of the selective pressures that would be operating, the difficulties such changes would cause for the organism, the expected time scale over which the changes would be expected to occur, the likely population sizes available in the relevant ancestral species at each step, other potential ways to solve the problem which might interfere, and much more.(For those who are wondering what that "much more" might even be, let me offer another prognostication: if an IC system was shown to have evolved to the level of detail demanded by Behe, his next step back would be to demand an account that each individual mutation was truly random with regard to fitness, as opposed as "poofed in" by the Designer. The ID goalposts have well-oiled wheels.) But does this demand even make sense with respect to IC? It is worth remembering that IC, the ID advocates hoped, was supposed to be the silver bullet that takes out "Darwinism", the one answering Darwin's own challenge: If it could be demonstrated that any complex organ existed, which could not possibly have been formed by numerous, successive, slight modifications, my theory would absolutely break down. There is, Behe and the ID advocates argued, something intrinsically special about IC, that makes it particularly impervious to Darwinian explanations. An irreducibly complex system, if there is such a thing, would be a powerful challenge to Darwinian evolution. Indeed, Dr. Behe has no problem at all with Darwinian explanations as they apply to other, not irreducibly complex systems. For instance, Behe accepts that hemoglobin (the protein complex that carries oxygen in red blood cells) evolved from myoglobin (the protein that stores oxygen within muscle fibers). Here's what he said about this: The question is, if we assume that we already have an oxygen-binding protein like myoglobin, can we infer intelligent design from the function of hemoglobin? The case for design is weak. The starting point, myoglobin, can already bind oxygen. The behavior of hemoglobin can be achieved by a rather simple modification of the behavior of myoglobin, and the individual proteins of hemoglobin strongly resemble myoglobin. So although hemoglobin can be thought of a system with interacting parts, the interaction does nothing much that is clearly beyond the individual components of the system.Behe even goes on to compare hemoglobin to the "man in the moon": suggestive of design, but almost certainly an illusion. But wait a minute: does Behe have in hand the list of mutations that occurred on the path from myoglobin to hemoglobin? Does he have "a detailed account of the selective pressures that would be operating, the difficulties such changes would cause for the organism, the expected time scale over which the changes would be expected to occur, the likely population sizes available in the relevant ancestral species at each step, other potential ways to solve the problem which might interfere, and much more"? You can try asking him, but I doubt it. The reason why Behe has no qualms with the evolution of the hemoglobin system is that it makes sense. The available evidence for precursors, intermediates and their functions, partial as it is, is sufficient to conclude that known, well-characterized evolutionary processes were responsible, as opposed to supernatural intervention. It really doesn't matter what every single amino acid substitution did in the long-extinct critters that evolved hemoglobins: only someone incompetent of biology, or an unrepentant Creationist, would require that level of detail. Behe knows that's absurd. That Dr. Behe asks for such an unnecessary level of detail for the evolution of the immune system (or any other IC system) carries two implications. First, it essentially reduces the concept of "irreducible complexity" to just a special case of evolution incredulity in general. Arguments from incredulity never go away (see Behe's "and much more", discussed above). In the case of evolution, we cannot have a mutation-by-mutation, selective-step-by-selective-step of pretty much anything, because the evidence cannot work that way, just like the evidence for plate tectonics can never be an inch-by-inch historical account of all the relevant forces involved in the motion of continents after the break-up of Pangea, or in the rise of the Himalayas. Even when we can make a very strong inference of selective effects on a protein's evolution (like in this case), we are still stuck with a level of detail that cannot compare to the absurd detail Behe is demanding. By insisting on a degree of evidence for IC systems' evolution that even evolutionary accounts of much simpler systems cannot provide, Dr. Behe has therefore effectively conceded that the concept of "irreducible complexity" is utterly meaningless: there is nothing special about IC systems, they just look fancy. In other words, it is not the "multiple, necessary, interacting parts" that make IC something that supposedly resists darwinian interpretations - it is amino acids, selective pressures, effective population sizes, like every other protein. Sic transit... To get a sense of how silly the argument actually becomes, consider the following. Below is an alignment of the simple, 30-amino acid B peptide of insulin in a few species. Many positions match, some do not.  With a little luck and hard work, we may be able to sample enough organisms to have, at least for some branches, a real mutation-by-mutation account of the evolution of peptide B. But no matter how we try, we will never have "a detailed account of the selective pressures that would be operating, the difficulties such changes would cause for the organism, the expected time scale over which the changes would be expected to occur, the likely population sizes available in the relevant ancestral species at each step, other potential ways to solve the problem which might interfere, and much more". Why is insulin peptide B less of a challenge for Darwinian evolution than the adaptive immune system? Behe himself had summarized in his book what he saw as the insurmountable problem of immune system evolution - not amino acids and selective forces, but: In the absence of the machine [RAG1/RAG2], the parts [V, D and J gene segments] never get cut out and joined. In the absence of the signals [RSSs], it's like expecting a machine that's randomly cutting paper to make a paper doll. And, of course, in the absence of the message for the antibody itself, the other components would be pointless.This is the "problem" the current data largely address: despite Behe's disbelief, there was a simple way that machine could be put together, by integrating a RAG-bearing transposon (which we now know exists) into an immunoglobulin-like immune receptor already under selection for diversity (which we now know exists). This single event, which bypasses all of Behe's objections above, is actually no more complex than the transition from a monomeric myoglobin to an allosteric hemoglobin complex ("allosteric" is just a technical word for a protein that works by changing its shape). In fact, arguably it's simpler. But rather than admitting he was wrong, that the evidence for evolution of the adaptive immune system is solid, and strengthening by the day, Behe has chosen instead to sacrifice whatever significance IC ever had. He ate his own child, to survive another day. The second issue with Behe's argument goes back to my original Calvin and Hobbes post. In it, I was not trying to make the point that the study of the evolutionary origin of the immune system is over. Indeed, I said that thankfully there is much more to be learned. My point was to compare the lively and steadily progressing field of evolutionary immunology, in Calvin and Hobbes' box, to the stale air inside the IC cabinet, in which all efforts are directed at keeping the door tightly shut. This really highlights the difference between the ID view of science, and what science actually is. ID is about absolute philosophical claims -- it does not, cannot cope with the fact that science is a process. As a political movement, ID has no time to let science take its course -- it must provide an ideologically satisfying answer right away, for its fund-raisers and activists, and defend it to the end. That is why scientists put their efforts into collecting data bit by bit, and ID advocates put theirs in revising definitions and raising the evidence bar to protect their claims from the new scientific data. Even Behe now behaves more like a spin doctor than a scientist. Consider this: in his post, Behe repeats once again the canard that Russ Doolittle made a mistake referring to clotting factor-deficient mice a few years back (an accusation which was nicely debunked by Ian Musgrave right here on the Thumb). I am quite sure people have pointed out to Behe that his claim is false before. In fact, since we know ID advocates eagerly read the Thumb (it took Behe only 24 hours to respond to my previous post!), I doubt that Behe was unaware of Ian's argument as he penned his latest reply. Assuming Behe now will likely read this post, can we expect him to cease propagating this falsehood? We'll see. Finally, Behe states that Orr and I "seem to think that because Darwinists' fantastic claims are very difficult to support in a convincing fashion, they should be given a pass". That's simply ludicrous: just my own post described a decade worth of hard-earned experimental results (and that's just the tip of the iceberg) from dozens of scientists, published in the very best scientific journals, supporting an evolutionary hypothesis that Behe had embarrassingly dismissed without a thought. Compare this level of effort and accomplishment to that of Behe's and his fellow ID advocates': in the same decade, they have put out not a single iota of a positive result for ID, while the Discovery Institute was throwing away Ahmanson's millions at school board challenges and PR campaigns hailing the upcoming scientific revolution. I'll leave it to others to judge whether Behe's words are more arrogant or ignorant. The real question to consider is: who is asking to be given a pass for "fantastic claims" here, those who are collecting data to support their hypotheses, or those who are running away from them? Originally posted at The Panda's Thumb. |